Publications
For a full list see Google Scholar.

Human pluripotent stem cells and tissue-resident fetal and adult stem cells can generate endoderm-derived epithelial organoids that recapitulate key aspects of human development and physiology. In this study, we integrated single-cell transcriptomic data from 218 samples, encompassing organoids and related in vitro models of diverse endodermal tissues, to establish an initial human endoderm-derived organoid cell atlas. The atlas comprises nearly one million cells across a wide range of conditions, data sources, and experimental protocols. By systematically comparing cell types and states between organoid models and primary tissues, we harmonized cell annotations through reference-based mapping. Focusing on the intestine and lung, we demonstrate how the atlas can be used to benchmark new protocols and assess perturbations and disease models. This resource centralizes diverse datasets and provides a foundation for evaluating organoid fidelity, characterizing diseased states, and accelerating protocol development.
Xu, Q.*,#, Halle, L.*, …, Treutlein B.#, Theis, F.#, Camp, JG.#
Highlight in Roche and Human Cell Atlas
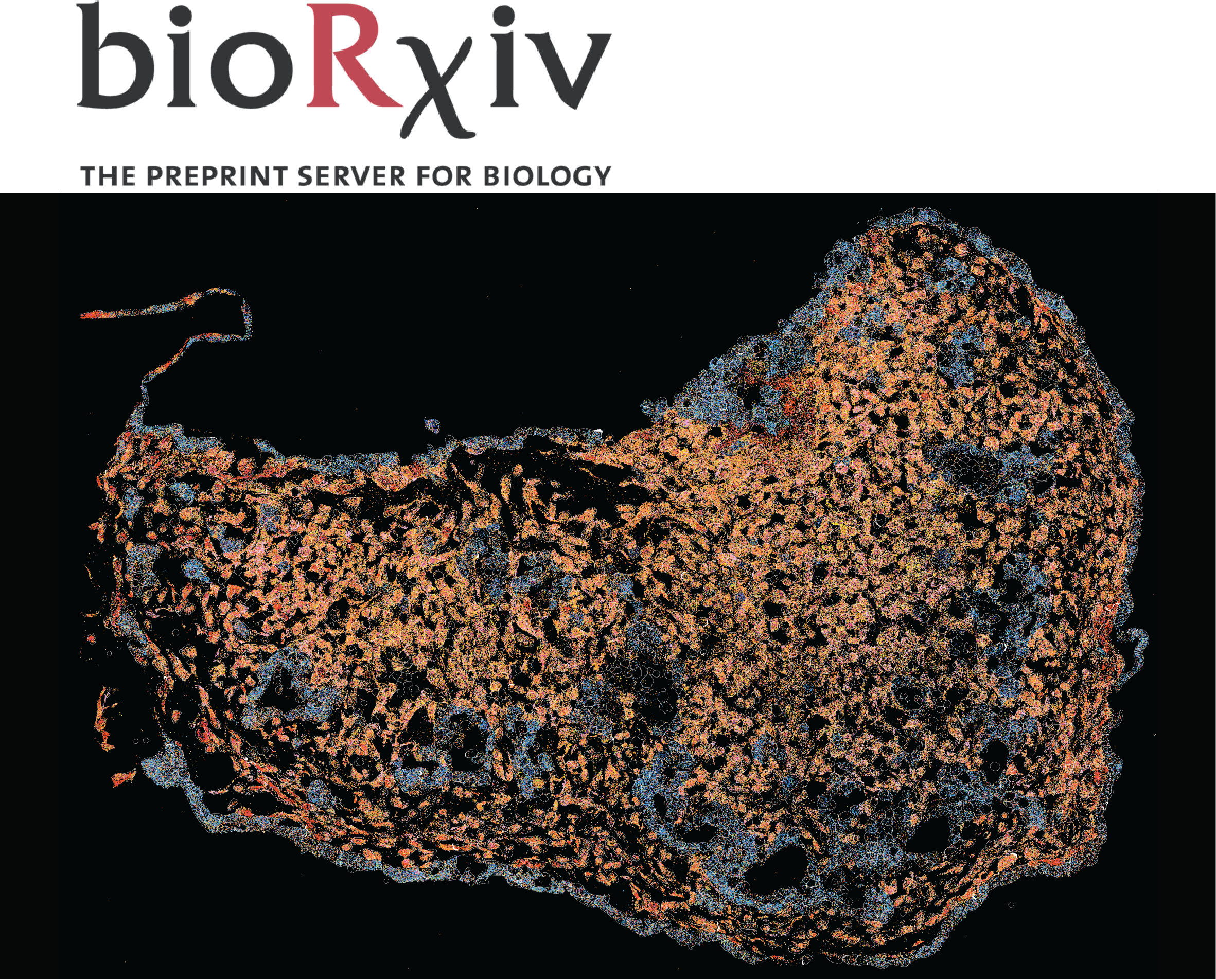
Pancreatic ductal adenocarcinoma (PDAC) is marked by a dense, fibroblast-rich stroma that actively shapes the tumor microenvironment and contributes to its aggressive clinical behavior. To define conserved PDAC cell states and uncover therapeutic vulnerabilities, we integrated single-cell transcriptomic data from 200 patient samples to construct a comprehensive PDAC atlas, identifying recurrent cancer cell and cancer-associated fibroblast (CAF) states, gene expression programs, and ligand–receptor interactions. We then developed modular patient-derived tumoroids incorporating both cancer cells and CAFs that recapitulate key features of ductal architecture and desmoplastic stroma. Single-cell and spatial transcriptomic analyses confirmed preservation of essential cellular states and signaling networks in vitro. Using this system, we identified Syndecan-1 (SDC1) as a CAF-responsive cancer cell receptor associated with poor patient survival, and demonstrated that functional blockade of SDC1 suppresses tumor growth in tumoroids. This work establishes an integrated atlas-to-organoid framework for dissecting cancer–stroma interactions and identifying actionable therapeutic targets.
Xu, Q.*, Okuda, R.*, Gjeta, B.*, …, Treutlein B.#, Camp, JG.#
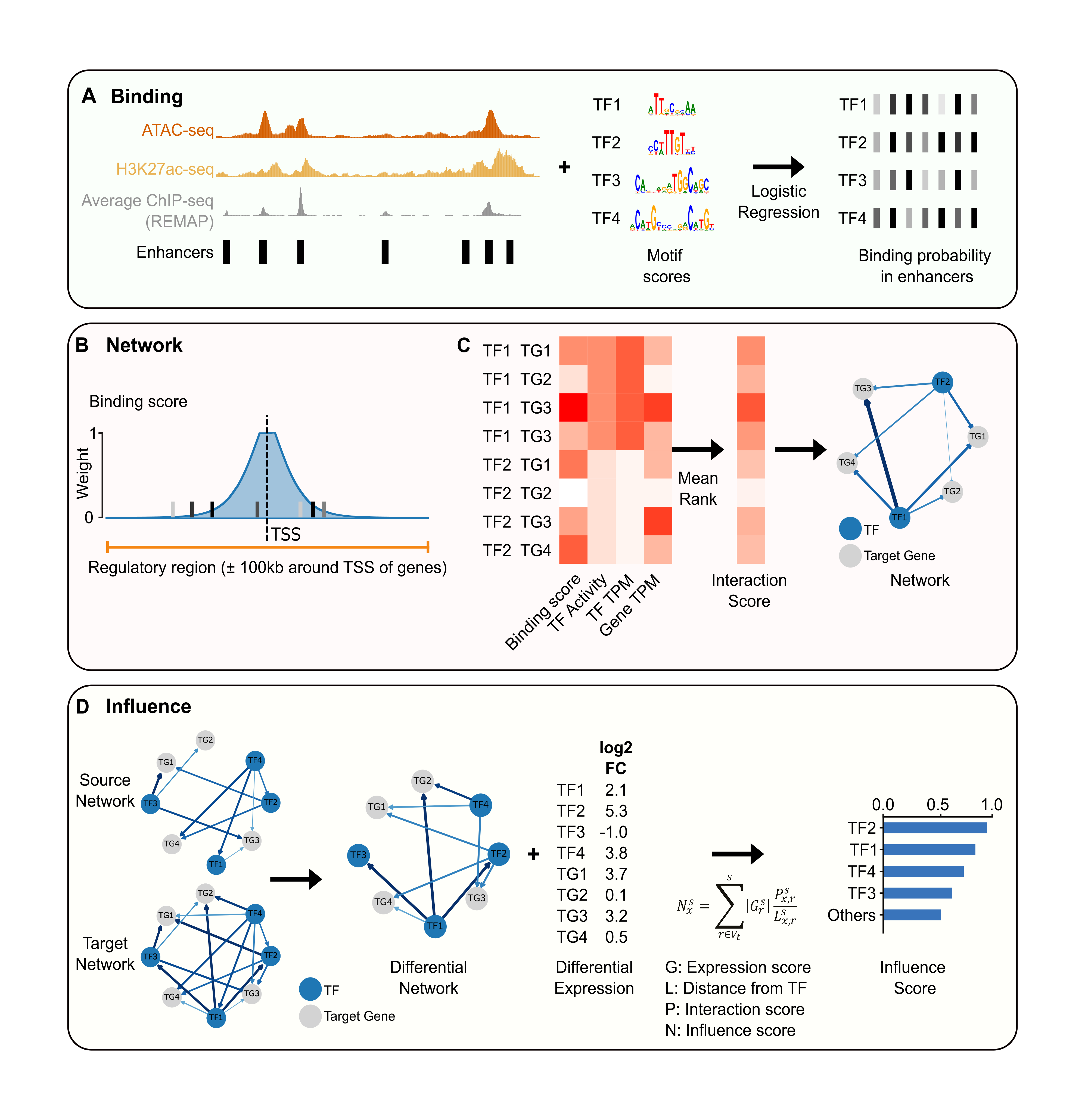
ANANSE introduces an enhancer-based framework for inferring gene regulatory networks and identifying key transcription factors that drive cell fate transitions. By integrating enhancer activity with transcription factor binding and gene expression, ANANSE captures regulatory mechanisms that are missed by promoter-centric approaches. The method consistently outperforms existing tools in predicting transcription factors capable of inducing trans-differentiation and provides mechanistic insights into transcriptional control during development and differentiation. ANANSE is open source, broadly applicable to custom datasets, and enables systematic prioritization of candidate regulators for experimental validation.
Xu, Q., Georgiou, G., Frölich, S., van der Sande, M., Veenstra, G.J.C., Zhou, H.#, and van Heeringen, S.J.#
Available at GitHub
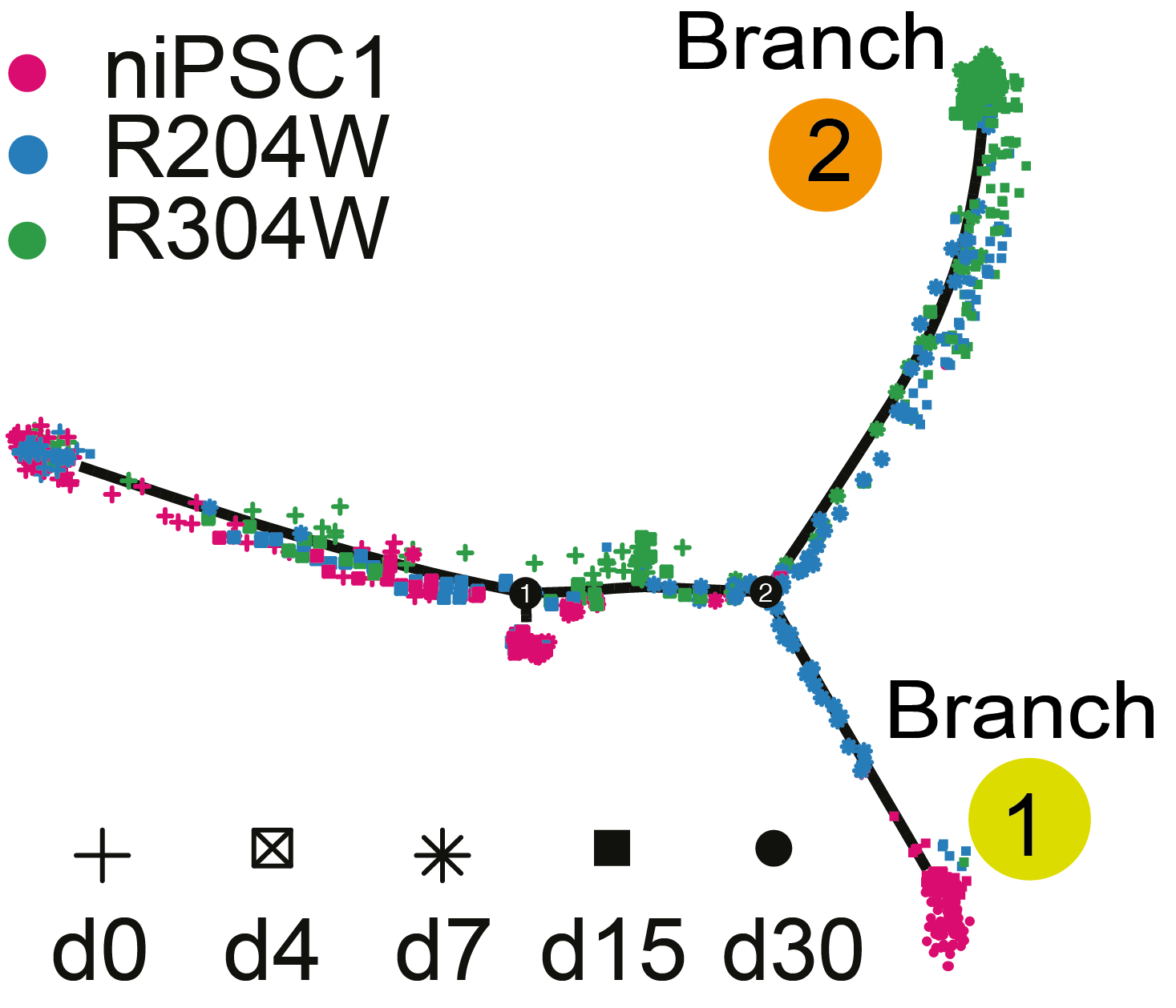
Using the transcription factor TP63 as a model, we investigated how transcription factor function and mutation influence cell fate determination and disease mechanisms. We studied human induced pluripotent stem cells (iPSCs) derived from EEC syndrome patients carrying DNA-binding domain (DBD) mutations in TP63, and analyzed transcriptomic changes during their differentiation into epidermal cells. We found that TP63 DBD mutations impaired proper epidermal fate specification and led to aberrant activation of mesodermal programs. Notably, inhibition of mesodermal induction pathways partially rescued epidermal commitment. This work provides mechanistic insights into TP63-associated disease and suggests potential therapeutic strategies for correcting cell fate defects caused by transcription factor mutations.
Soares, E.*, Xu, Q.*, Li, Q.*, Qu, J., Zheng, Y., Raeven, H.H., Brandao, K.O., Petit, I., van den Akker, W.M., van Heeringen, S.J., Aberdam, D., Tang, F.#, and Zhou, H.#
Proceedings of the National Academy of Sciences (2019)
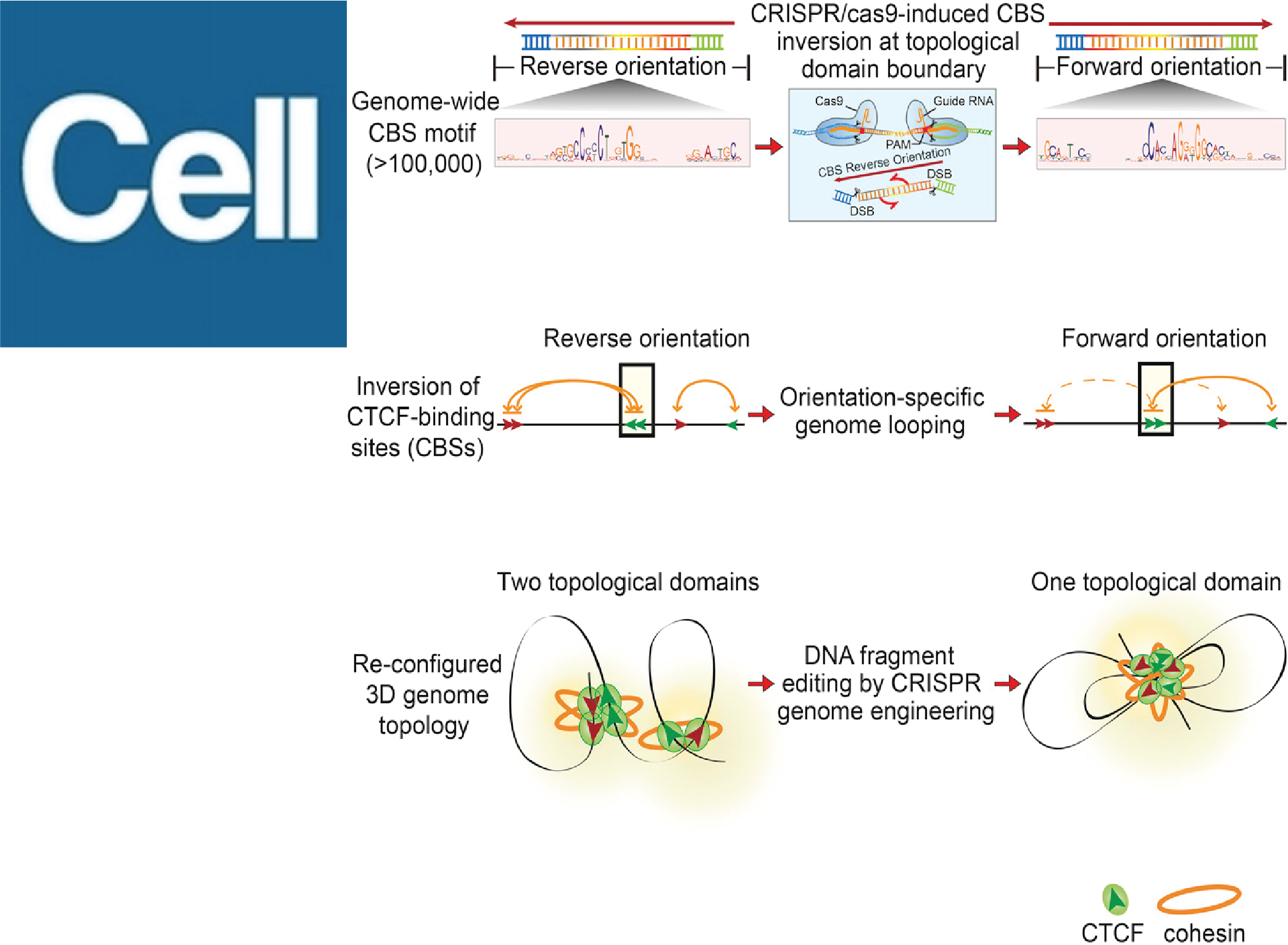
The architectural protein CTCF plays a central role in shaping the functional output of the genome. In this study, we demonstrate that the orientation of CTCF binding sites constrains their choice of interacting partners, establishing an orientation-dependent code that predicts higher-order three-dimensional genome organization. We further propose a DNA loop extrusion model to explain how CTCF orientation governs specific chromatin loop formation.
Guo, Y.*, Xu, Q.*, Canzio, D., Shou, J., Li, J., Gorkin, D. U., Jung, I., Wu, H., Zhai, Y., Tang, Y., Lu, Y., Wu, Y., Jia, Z., Li, W., Zhang, M.Q., Ren, B., Krainer, A.R., Maniatis, T.#, and Wu, Q.#
Highlight in Cell and Nature Reviews Molecular Cell Biology